Molecular dynamics
We consider molecular dynamics (MD) as
an approach to obtain:
1.
Thermodynamic properties (energy, heat capacity etc.)
2. Dynamic information (diffusion
coefficient, dielectric functions, correlated motion etc.)
MD is a classical approach based on
Newton’s equations of motion. Each atom
in a structure is assigned parameters that describe
both bonding and non-bonding interactions.
The parameters are shown graphically below. The potential energy of the system is given by these terms.
The kinetic energy is given in the form of a
Maxwell-Boltzmann distribution of velocities.
In
order to propagate a molecular system using a force field there are three
typical stages.
1.
Minimization
2.
Equilibration
3.
Dynamics

Figure 1. Graphical representation of the terms that
contribute to a force field.
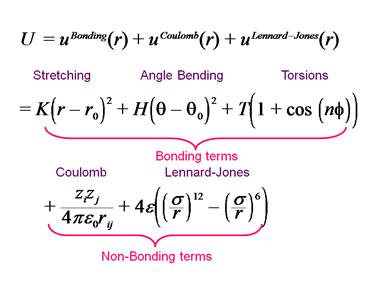
Figure 2. Mathematical form of terms that contribute
to a force field.
For macromolecules (peptides,
oligonucleotides, and oligosaccharides) the
conformations can involve many different possible states of nearly the same
energy. The interconversion involves
thermal motion. Crossing from one
conformation to another involves transit over a barrier. If kBT
is greater than the barrier height then this will occur with high
probability. If kBT is less than the barrier height the
system will be confined to a smaller region of conformational space.
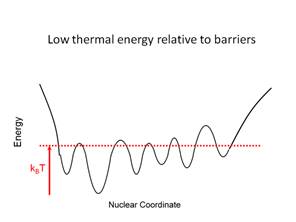
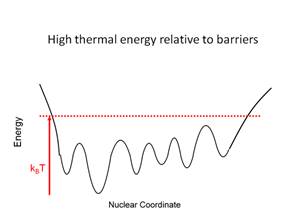
Figure
3. Graphical representation of the
relative magnitude of thermal energy and energy barriers in the force field.